这篇是《Exploring Protein Structure: Principles and Practice》这本书的练习解答。
纯粹是练习记录。这是Exercise 2的部分。
本系列是《Exploring Protein Structure: Principles and Practice》这本书的练习解答。标题的EPSPP是书的题目的缩写。考虑到第一章没有练习,所以从Exercises-2开始。
《Exploring Protein Structure: Principles and Practice》是Springer在2018年出的一本教材,属于Learning Materials in Biosciences系列。本书是作者Tim Skern多年讲授蛋白质结构与功能课程的一个产物。对任何想从事生物医学相关的小伙伴而言,这都是一本绝佳的入门书。没有枯燥的概念,作者通过PDB数据库和PyMOL软件,交互地讲解蛋白质结构和功能的各个知识点。通过对PyMOL的学习,读者会对蛋白质这个奇异的物质逐步了解理解,进而爱上它(相信我)。考虑到这并不是一个本书的笔记,因此我就在这里停下行文。下面开始练习解答的记录。(考虑到原文是英文,我这里会偷懒使用英文记录题目,不定期更新)
2.6.1
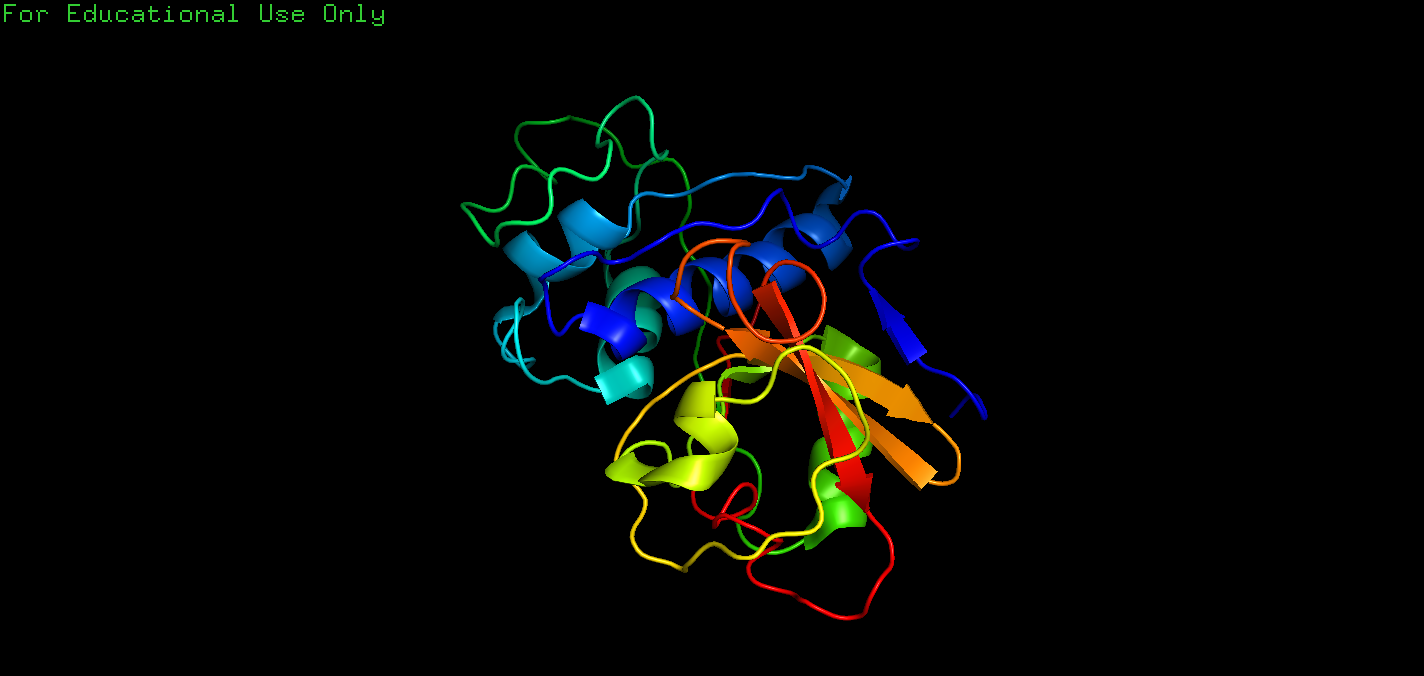
Q: The first exercise asks you to draw the image of a humble protein that given tremendous insight into protein structure and function over tha last 40 years. The protein is the enzyme lysozyme from the T4 bacteriophage. Elementary to produce in bacteria, straightforward to crystallize, and simple to measure, it was one of the first proteins whose structure was solved as well as being one of the first to be subjected to site-directed mutagenesis(定点诱变). Mutation of T4 lysozyme has enormously magnified our knowledge of protein structures and enzyme mechanisms. We will see in Sect9.2 that this extremely stable protein is presently proving extremely useful in helping to crystallize difficult proteins. Figure 2.7 illustrates this protein using the PDB entry 2lzm. Can you make this drawing? The command for the spectrum is “util.rainbow”.
A: The code is:
1 | fetch 2lzm |
2.6.2
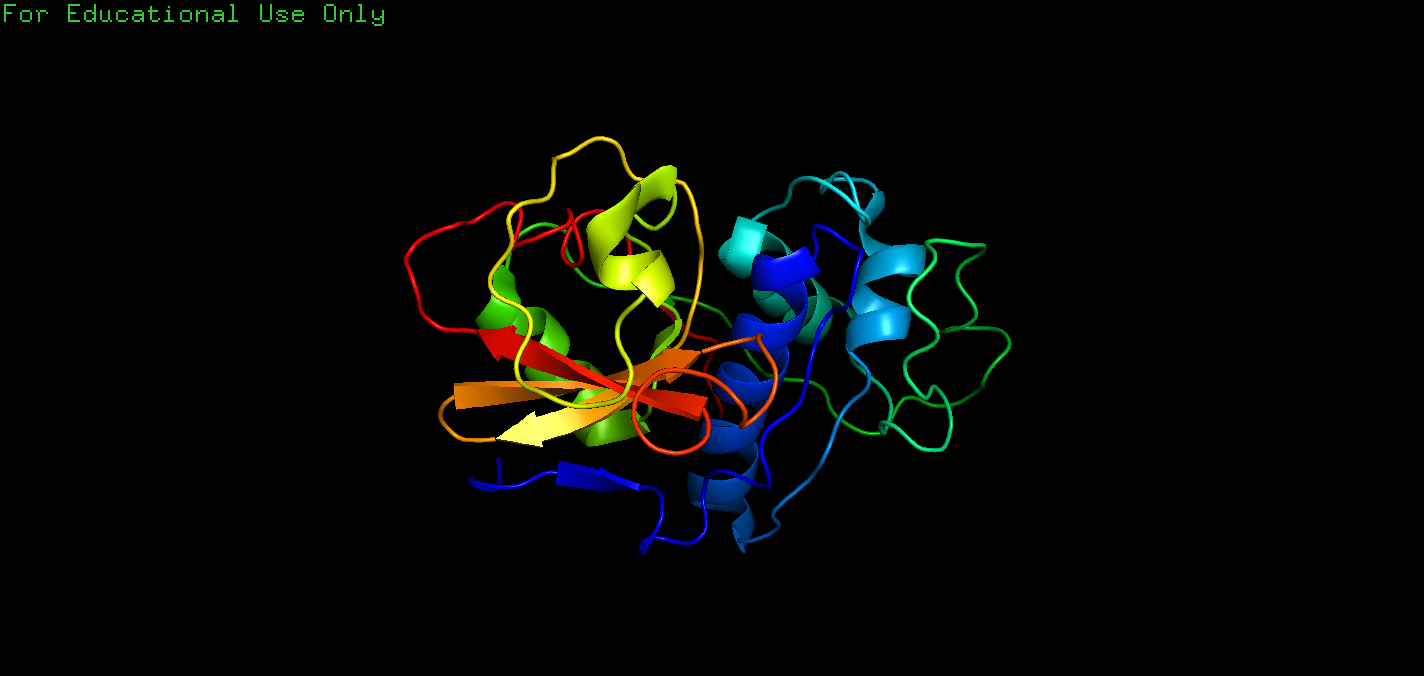
Q: The second exercise involves a search of the PDB for structures of the protease papain, a molecule that we will explore in the next chapter. Enter papain into the search box of PDB, and you will see around 200 hits. Papain is a prototype for a group of proteases, so it often appears in the titles of the entries because of a mechanistic and structural resemblance. To find the structure that we will use in Chap.3, we need to refine the query. At the top of the hits for papain is the button “refine search”. Clicking this button gives you the opportunity to optimize your original search by taking you to the “Advanced Search Interface”. This tells us that entering papain into the search box on the home page initiates a “Text Search” that looks through every field in every entry. To refine the search, click on the little arrows at the right side of “Text Search”, scroll down to the group of fields “Structure Annotation”, and click on “Structure Title”. Enter papain in the search box. Then, in the “Choose a Query Type” box underneath, click on the small arrows, and scroll down to “Author Name” in the “Deposition” field and enter “Drenth”. Before you submit the query, let us just look at the feature “Result Count” at the right of each query box. If you click this, you will see how many entries for this term are present in the PDB; in October 2017, this was 59 for papain and 17 for Drenth. This feature is very useful to see if you have too many hits or none at all! Now click “Submit query”, which should give six entries; that is, of the 17 structures deposited by Drenth, 6 contain papain in the structure title. The first hit, 9pap (Kamphuis et al. 1984), will be analyzed in Sect.3.3. Note that you cannot view the file in PDB format from the search results. You have to click on the entry (e.g., 9pap) and then download from the summary page. Downloading the files from the search results gives a file in the “cif” format, which stands for “Crystallographic Information File” which cannot be read by PyMOL(NOW IT CAN), unless you clicked the option to read such files on installation. Try to draw a cartoon of papain using the coordinates in the entry 9pap using the commands you have learnt already in this chapter. You can then compare it with Fig.3.3 in Chap. 3.
A: The code is:
1 | fetch 9pap |
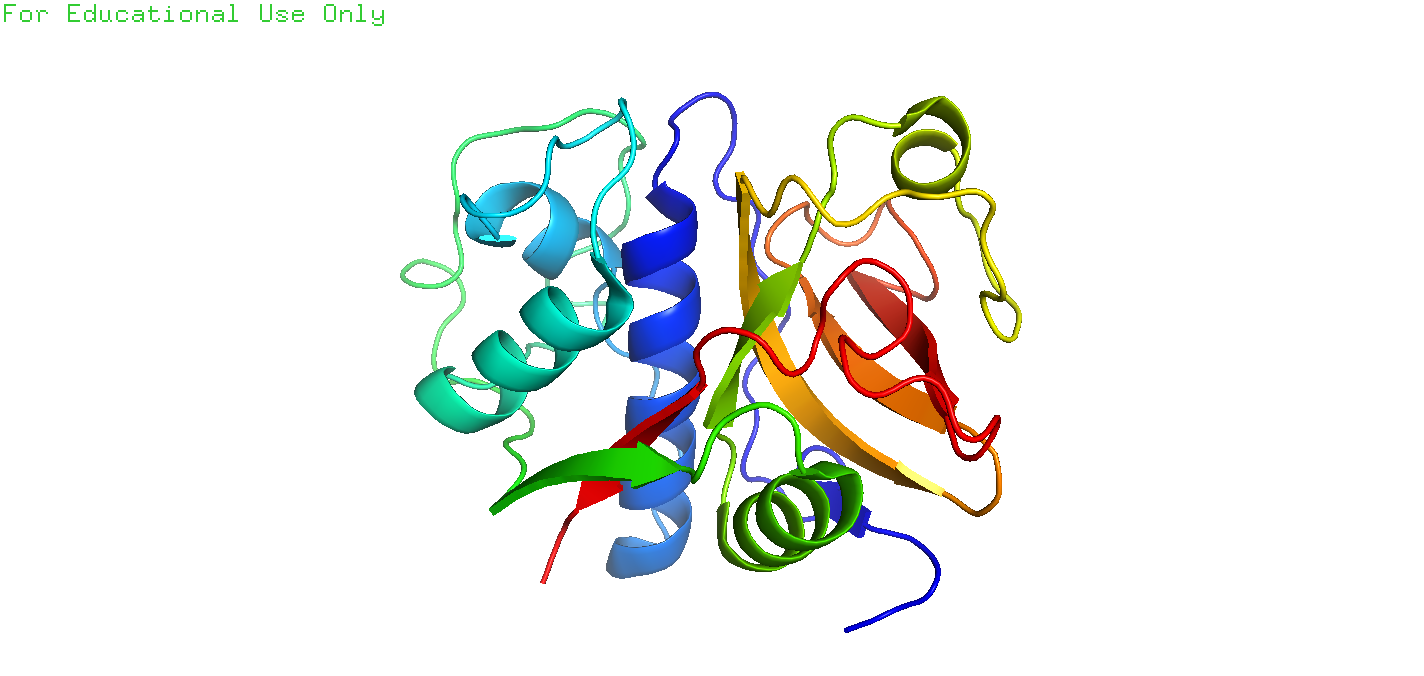
2.6.3
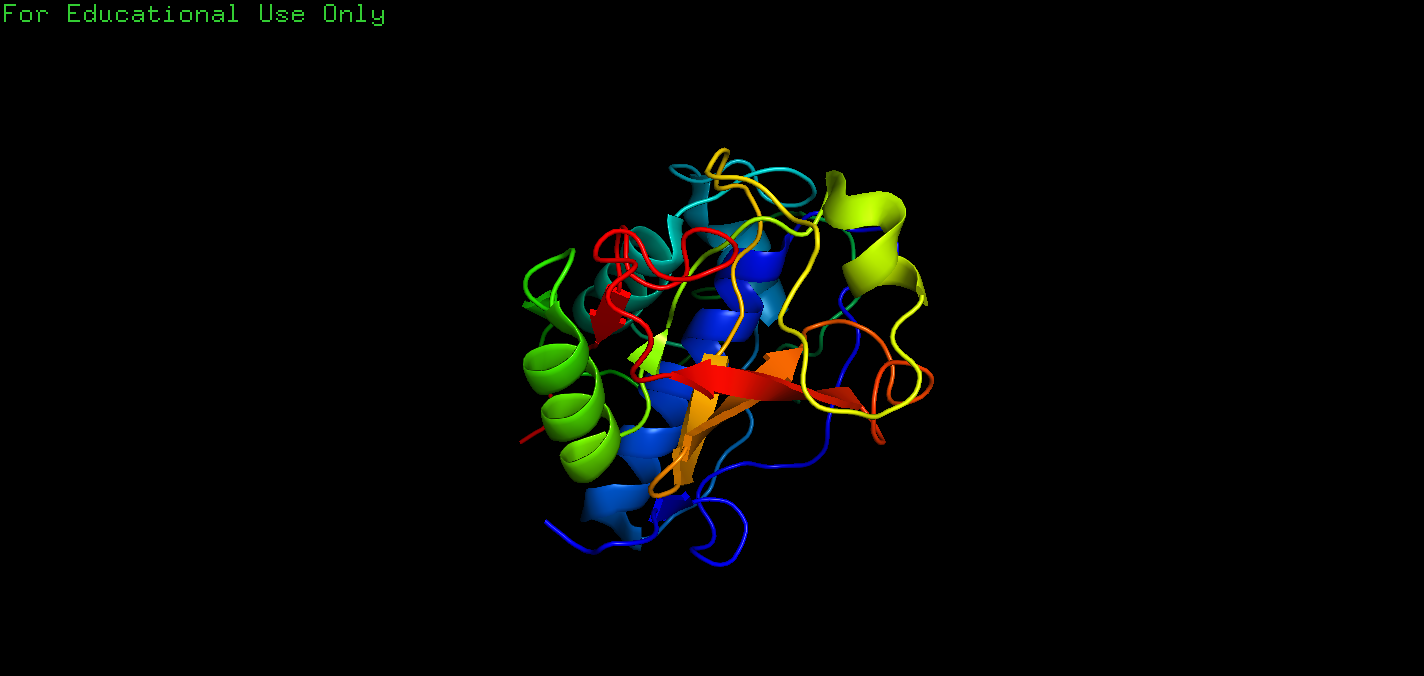
Q: The other hits in the search for papain and Drenth in Exercise 2.6.2 are all papain structures determined during a study of the interaction of papain with chloromethylketone inhibitors(氯甲基酮抑制剂) (Drenth et al. 1976). Look at the entry 4pad in PyMOL. Visualize the inhibitor TCK (Tosyl-L-lysine chloromethyl ketone) in the same way as for 2NC in Box 2.3. How does the binding of the inhibitor TCK differ from that of 2NC? We will also examine this interaction in Exercise 6.13.1 in Chap. 6 and compare the binding of the two proteases in more detail.
A: The code is:
1 | fetch 4pad |
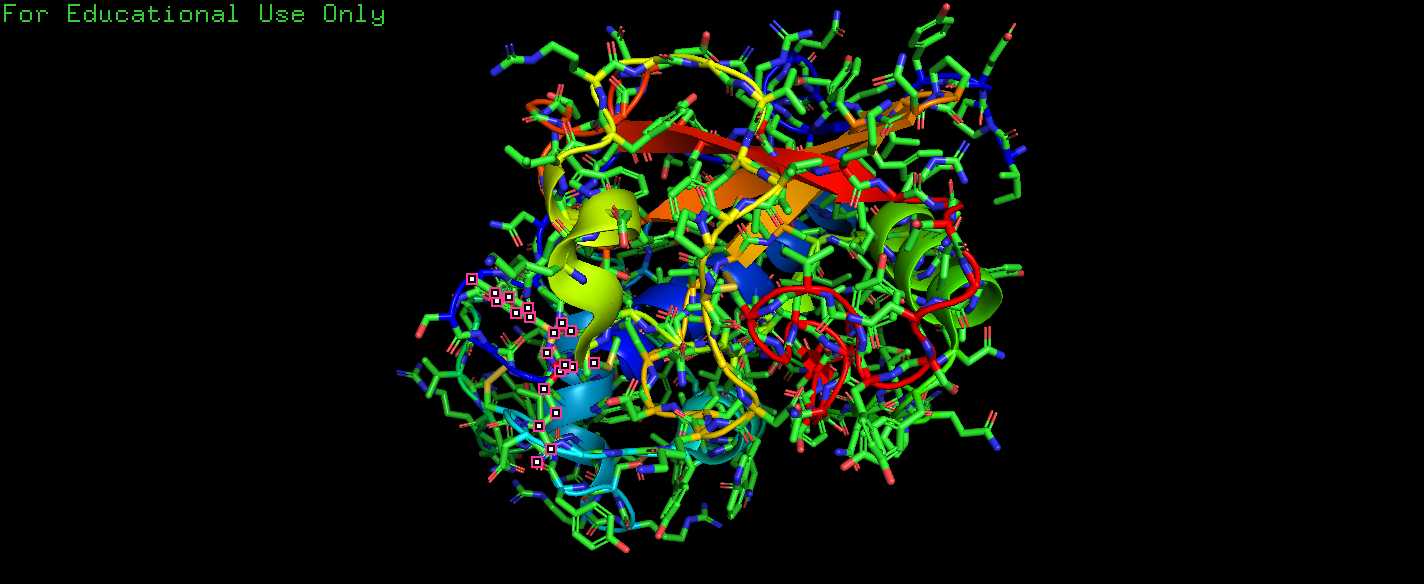
2.6.4
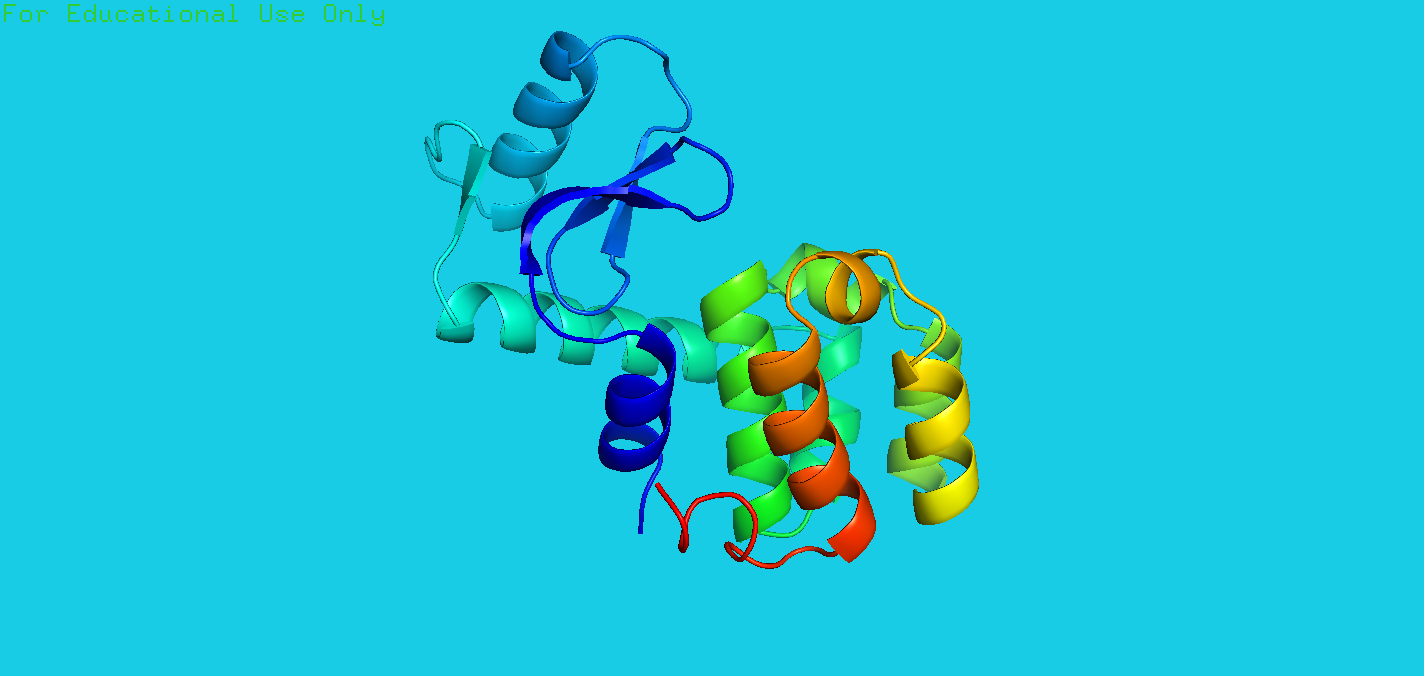
Q: Choose any one of the molecules that you drew in this chapter, and try to imagine how the protein structure will change when you rotate it by 90° about each of the three axes. This will help you to better visualize the proteins in three dimensions.
A: The code is:
1 | fetch 4pad |